Computational Biology
Recent news:
Professors Paul Adams, Adam Arkin, Patrick Hsu, and Jay Keasling have been recognized in the “2025 Highly Cited Researchers” list, meaning their work ranks in the top 1% of citations for their field and publication year in Clarivate’s Web of Science citation index
Adjunct Professor Taner Sen and his colleagues at the USDA and beyond have assembled and annotated the genomes of 33 wild and domesticated oat lines, along with an atlas of gene expression across 23 of these lines, which will enable future efforts to produce even more hardy and productive strains of the popular grain.
Professor Liana Lareau is recognized for her revolutionary approach to treat retinitis pigmentosa and other dominant genetic diseases by combining CRISPR prime editing with machine learning.
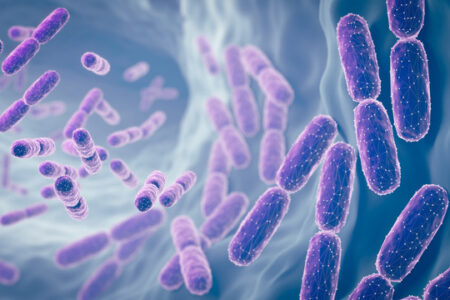
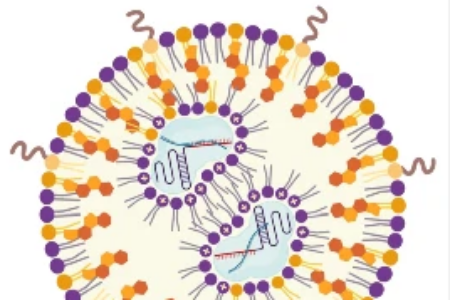
Computational modeling from Mofrad Lab gives us a peek inside these important microbial communities.
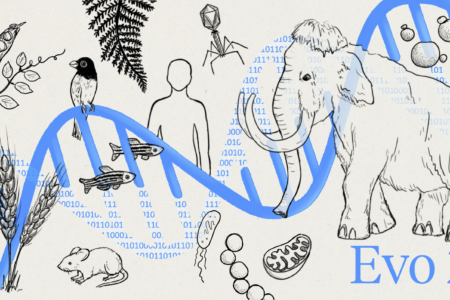
Research led by Professor Patrick Hsu has produced Evo 2, the largest AI model in biology to date, which can accurately predict the effects of all types of genetic mutations.
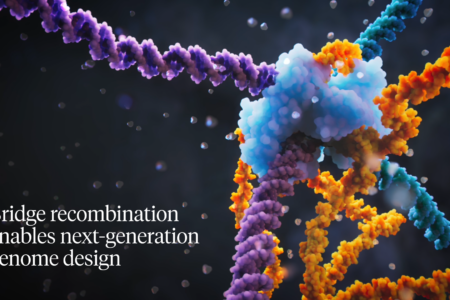
Patrick Hsu’s breakthrough discovery of bridge RNA gene editing tools is discussed on WebMD, with contributions from BioE alumnus Connor Tou.
Murthy lab and UC Davis have developed a unique mRNA delivery method for in-utero gene editing for neurodevelopmental conditions.
Niren Murthy et al. have developed a more stable version of the Cas9 enzyme to improve delivery of CRISPR-Cas9 ribonucleoproteins (RNPs) for in vivo gene editing.
Professor Leah Guthrie works to understand how the microbiome metabolites and proteins communicate with our human cells to influence our physiology and pathophysiology. Learn more about Guthrie in this interview with QB3.